What is host cell protein?
Host cell protein (HCP) is the largest protein contaminant in biopharmaceuticals, derived from recombinant DNA technology. Most biotherapeutics use recombinant DNA technology to express target protein drugs using preferred host cells. Since host cells also express a large amount of protein, recombinant protein drugs will be contaminated by HCPs. Even after complicated purification steps, low concentrations (1-100 ppm) of HCP may remain in the final biopharmaceutical product.
The residual HCP content in biological products is usually a critical quality attribute (CQA) of the product. The remaining host cell protein as a foreign protein may trigger immune response of the body to varying degrees, and even cause anaphylactic shock.
In addition to safety issues, the presence of HCPs can also affect product quality. For example, HCPs can trigger the aggregation or fragmentation of therapeutic proteins. The protease activity in HCPs can also affect the protein composition in the culture supernatant, thereby affecting the subsequent protein purification process or the long-term storage stability of protein drugs.
In addition, HCPs may undergo post-translational modifications, making quantification and characterization more difficult.
HCP may affect the safety and efficacy of the product. Regulatory agencies such as the World Health Organization, the European Union, and the FDA require the identification and quantification of HCP of biopharmaceutical products to ensure safety.
How to analysis host cell protein?
Due to the wide linear dynamic range of protein concentration in protein drugs (covering four to five orders of magnitude), HCP detection is challenging. During the development and production of genetically engineered drugs, several methods can be used to detect HCPs and other impurities.
1. Enzyme-linked immunosorbent assay (ELISA)
Among the methods of HCPs analysis, anti-HCPs ELISA is considered the gold standard. This method (detects protein concentration in a range of 1~100 ppm) is suitable for product development and process control. However, unlike the conventional ELISA, the anti-HCPs ELISA does not detect a single target, and the total HCPs “antigen” is a complex protein group. The conventional HCPs antibody preparation method is to use host cells containing empty expression vectors to prepare antigens to immunize animals to obtain antiserum, and isolate and purify polyclonal antibodies for ELISA detection. When producing HCPs antibodies/standard reagents, it is necessary to ensure the specific production cell line and production process as well as the accurate quantification of its protein concentration. The universal HCP ELISA method developed using polyclonal antibodies produced against parental cell lysates or cell culture supernatants can detect most HCP species, but may not detect protein subsets specific to a particular manufacturing process.
At present, the design and verification of HCPs immunoassay face the following challenges:
1) The dynamic range is narrow (<100), the development cycle is long, the cost is high, and the risk of failure is high.
2) There are a wide variety of possible HCPs in genetic engineering drugs.
3) Generally, polyclonal antibody reagents are used for detection, which may lead to inaccurate results.
4) Different proteins have similar epitopes, resulting in false positives.
5) Hook effect. High abundance proteins lead to detection saturation.
6) Lack of quantitative criteria for perfect matching.
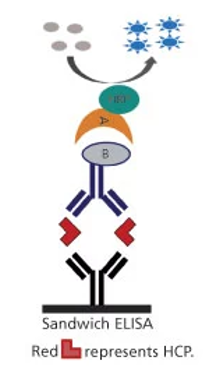
Schematic diagram of sandwich ELISA for HCP detection and measurement (Wang et al., 2015)
2. Two-dimensional gel electrophoresis (2-DE)
2-DE used with fluorescence staining is another commonly used HCP analysis method. 2-DE is often used in upstream or downstream process development and characterization. The principle of 2-DE is to separate different proteins according to the isoelectric point (first dimension), and also according to the size (second dimension). The spot analysis in the gel does not require immunoblotting, which avoids the membrane transfer step, and can separate trace HCPs impurities from the product. But this method can only perform semi-quantitative analysis, since its dynamic range is narrow (two to three orders of magnitude), and it needs to be combined with other techniques (such as mass spectrometry) to identify HCP.
3. Mass spectrometry (MS)
MS is increasingly used to detect HCPs in order to help identify possible risk factors and evaluate the purification process. Although ELISA is most commonly used to detect the total amount of HCPs, it cannot recognize HCPs outside the detection range of the ELISA kit. Compared with the traditional ELISA method, the mass spectrometry method has the advantages of short development time, specific confirmation and quantification of all HCPs, unbiased detection of all impurity proteins, high dynamic range (up to 6 orders of magnitude), and fast adjustment.
Universal UPLC/MS analysis can comprehensively identify and quantify HCP in biotherapeutic protein samples. This analytical method uses online two-dimensional liquid chromatography to separate peptides, and then uses a high-resolution, high-quality and accurate mass spectrometer for protein identification and quantification.
Parallel reaction monitoring (PRM) mass spectrometry quantitative technology can scan all the product ions produced by a parent ion, with high identification accuracy and quantitative throughput, and can accurately identify target HCPs peptides.
Reference
Wang F, Richardson D, Shameem M. Host-cell protein measurement and control. BioPharm Int, 2015, 28: 32-38.



